-
ChIP training
Part 1: Basics
Welcome to our training series on chromatin immunoprecipitation (ChIP). We'll show you ChIP basics and essential protocols before moving on to optimization, troubleshooting, and more advanced techniques.In Part 1 of our series on ChIP, we take you through the basics, background, and potential uses of this powerful technique. Before you start the protocol itself, familiarize yourself with the essentials behind the methodology.
Part 1 overview1.1 What is ChIP?
1.2 Chromatin structure and function
1.3 When to use ChIP
1.4 Advanced ChIP-based methods
1.1 What is ChIP?ChIP is a technique that allows you to analyze the localization of proteins binding to specific regions within the genome. ChIP, therefore, enables researchers to examine the interactions between epigenetic regulators and DNA in their natural context.
The central principle of ChIP is the use of an antibody targeting a protein of interest to pull down DNA regions with which it associates. You can selectively enrich for chromatin fractions using antibodies specific for a protein of interest (eg a transcription factor). After you have enriched for your protein of interest, the DNA regions in which the proteins are interacting can be isolated using a pull-down technique. The pull-downed DNA can then be sequenced (ChIP-seq) or used for qPCR (ChIP-PCR) to determine where in the genome your protein of interest is binding.
In this video, you’ll learn what the ChIP technique allows you to do.
With ChIP, researchers can identify specific genes and sequences where a protein of interest binds, across the entire genome, providing critical clues to their regulatory functions and mechanisms. By dissecting the temporal and spatial dynamics of protein-DNA interactions, ChIP provides insights into core biological processes and disease pathology.1.2 Chromatin structure and functionChromatin is comprised of histones and DNA. DNA wraps around eight core histones to form the basic chromatin unit, the nucleosome. Nucleosomes are strung together like beads on a string and packaged into higher-order chromatin architecture (Figure 1).
The function of chromatin is to efficiently package DNA into a small volume to fit into the nucleus of a cell and protect the DNA structure and sequence. Packaging DNA into chromatin allows for mitosis and meiosis, prevents chromosome breakage, and controls gene expression and DNA replication.
There are two main types of chromatin:
1) Heterochromatin – a tightly packed form of chromatin that can silence gene expression
2) Euchromatin – a less condensed form of chromatin that allows for active gene transcription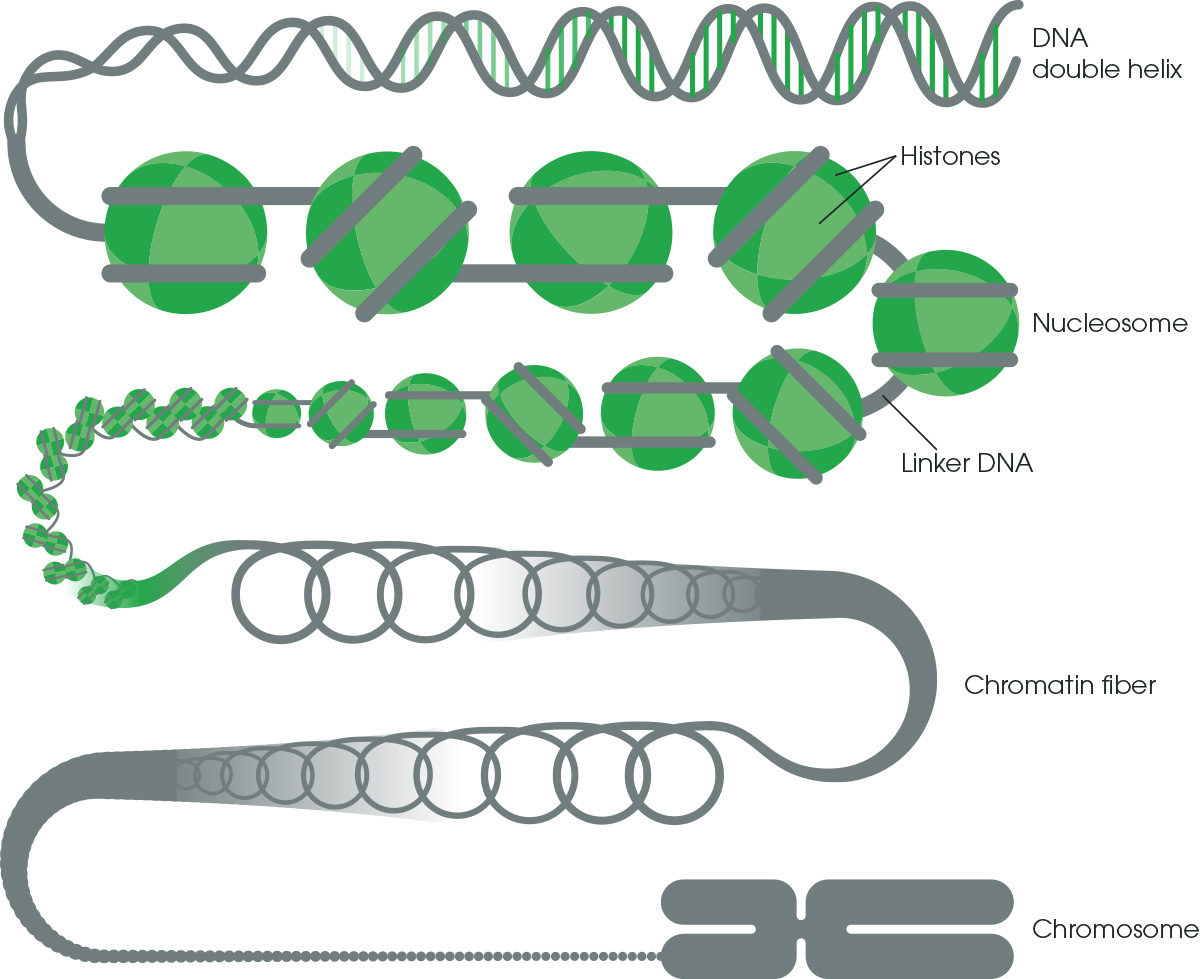
Figure 1. Chromatin structure. DNA winds around nucleosomes to form chromatin fiber and then chromosomes.
Chromatin is actively and dynamically remodeled to alter gene expression and cellular programming, for example, during different developmental stages or in response to particular stimuli. Large genomic regions may be silenced or activated, or nucleosomes may be unraveled to access specific genes and DNA sequences. Understanding which genomic regions are active or inactive across the genome in different cellular or disease states can help to identify critical pathways and associated genes.
Find out about the methods used to investigate chromatin accessibility and interactions here.1.3 When to use ChIPChIP is the technique of choice to study chromatin modifications. You can use ChIP to dissect the spatial and temporal dynamics of chromatin interaction with its associated factors. Most commonly, ChIP is used to detect the pattern of histone modifications and DNA-binding proteins such as transcription factors across the whole of the genome.
Histone modifications
Histone tails protruding from the nucleosome core can be targeted for modification. Different modifications will have different effects on chromatin conformation and gene expression. It is, therefore, very common to carry out ChIP for these modifications to obtain a clearer picture of the chromatin landscape surrounding your gene of interest. Certain histone modifications are also associated with different gene regulatory regions such as enhancers or promoters.
Histone H3 is the most commonly modified histone (see Figure 2); specifically, modifications include methylation (me) or acetylation (ac) of lysine (K) residues within histone H3. These are some of the most commonly studied histone modifications and their known functions.Activation
H3K4me1 – enhancers
H3K4me3 – promoters
H3K36me3 – gene bodies
H3K9ac – promoters, gene-rich regions
H3K27ac – satellite repeats, telomeres, pericentromeres, retrotransposons
Repression
H3K27me3 – enhancers, promoters
H3K9me3 – enhancers, promoters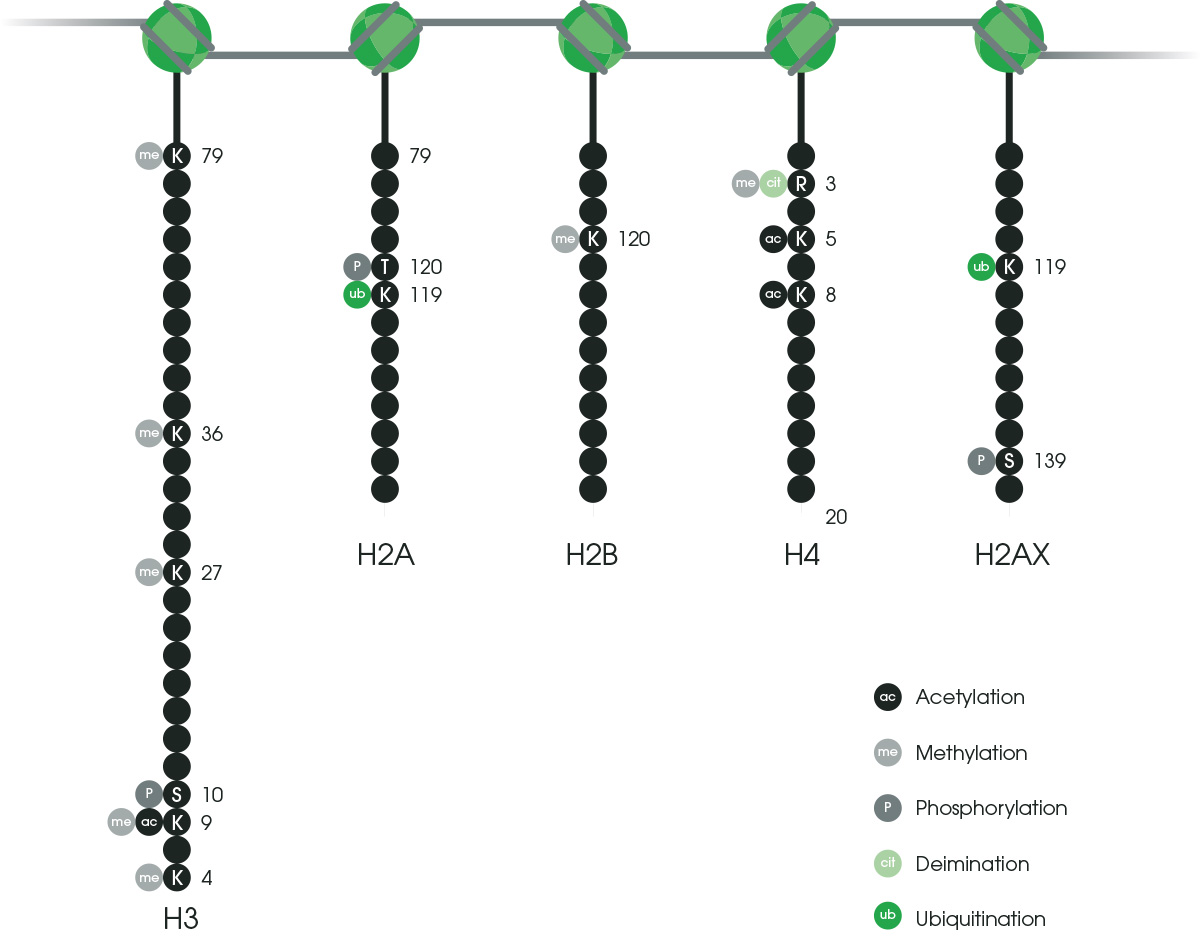
Figure 2. The most common histone modifications.
Together, these histone modifications (Figure 2) make up what is known as the histone code, which dictates the transcriptional state of the local genomic region. Examining histone modifications at a particular region, or across the genome, can reveal gene activation states, locations of promoters, enhancers, and other gene regulatory elements.
You can learn more about histone modifications in this guide.
Transcription factors
You can also use ChIP to analyze the location of transcription factors and other DNA-binding proteins. For example, the pluripotency transcription factors (eg Oct4 and Nanog) are known to bind and activate the transcription of the first zygotically expressed genes of early embryos. If you carry out ChIP for these factors using chromatin from early embryos, the DNA fragments you pull down will be these initial zygotically expressed genes. Another example using a DNA-binding protein is if you carry out ChIP for RNA polymerase II, you’ll find enrichment within actively transcribed genes, the promoter regions, and along the whole of the gene body. RNA polymerase II would not be detected at high levels in inactive genes.
In this video, we’ll have a closer look at a chromatin structure and its modifications and go through specific examples of how ChIP is used to study histone modifications and transcription factors.
1.4 Advanced ChIP-based methodsTraditional ChIP has acted as a springboard to develop novel adaptations, which often combine ChIP with other techniques to address diverse biological problems. For example, ChIP-loop, which uses restriction digestion of cross-linked chromatin and ligation of interacting fragments to determine the three-dimensional organization of the genome. ChIP can also be combined with epigenetic technologies such as bisulfite conversion (BisChIP-seq) to interrogate the interplay between histone modifications and DNA methylation.
Chromatin Interaction Analysis by Paired-End Tag sequencing (ChIA-PET) is a hybrid of ChIP and 3C techniques. ChIA-PET is a high-throughput version of ChIP-loop capable of detecting long-range chromatin interactions via a protein of interest across the genome.
Very slight modifications to traditional ChIP allow ChIP-exo to detect protein binding at single-nucleotide resolution. ChIP-exo uses restriction to digest and next-generation sequencing to determine genome-wide protein binding with high resolution. The resolution of ChIP-exo is greater than almost all other ChIP-based techniques.
You can find out more about the latest advanced ChIP-based techniques with this guide.Summary
You should now have a better idea of the ChIP basics. Here’s a quick summary:
• ChIP allows you to analyze the localization of proteins binding to specific regions within the genome
• By assessing protein-DNA interactions in their natural context, ChIP provides insights into core biological processes and disease pathology
• Chromatin is comprised of histones and DNA
• There are two main types of chromatin - heterochromatin and euchromatin - both having different effects on gene expression
• ChIP is used to detect histone modifications and DNA-binding proteins
• The basic ChIP protocol can be used in other more advanced techniques
In Part 2, we’ll introduce you to the cross-linking and native ChIP protocols as well as chromatin preparation from tissues and plant samples.
Start Part 2 now!